Publications (pubmed: reiter nj)
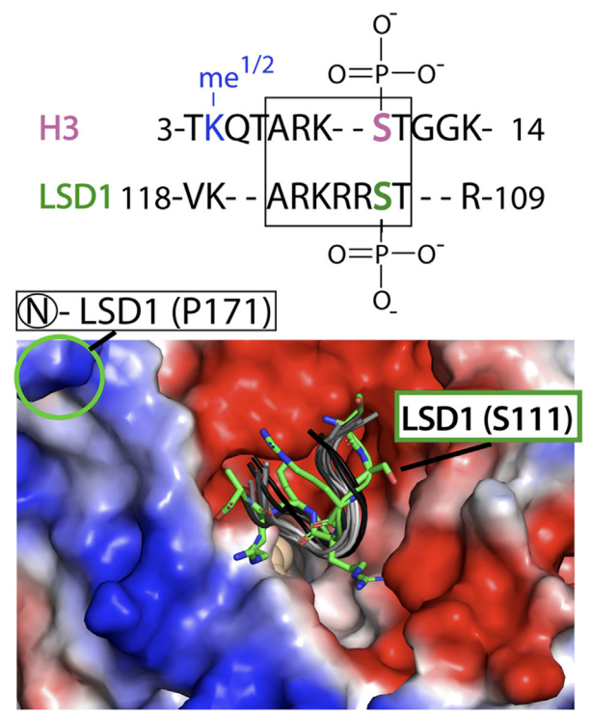
1. D. Senanayaka, D. Zeng, E. Deniz, I. K. Priyankara, J. Helmbreck, O Schneider, A. Mardikar, A. Uren, N. J. Reiter * “Autoregulatory mechanism of enzyme activity by the nuclear localization signal of lysine-specific demethylase-1.” Journal of Biological Chemistry, 2024, 300(9): 107607 - 10.1016/j.jbc.2024.107607. *Selected as an EDITOR’S PICK
2. D. Senanayaka, D. Zeng, E. Deniz, I. K. Priyankara, J. Helmbreck, O Schneider, A. Mardikar, A. Uren, N. J. Reiter * “Anticancer drugs of lysine specific histone demethylase-1 (LSD1) display variable inhibition on nucleosome substrates.” Biochemistry, (online 5/14/2024, doi: 10.1021/acs.biochem.4c00090).
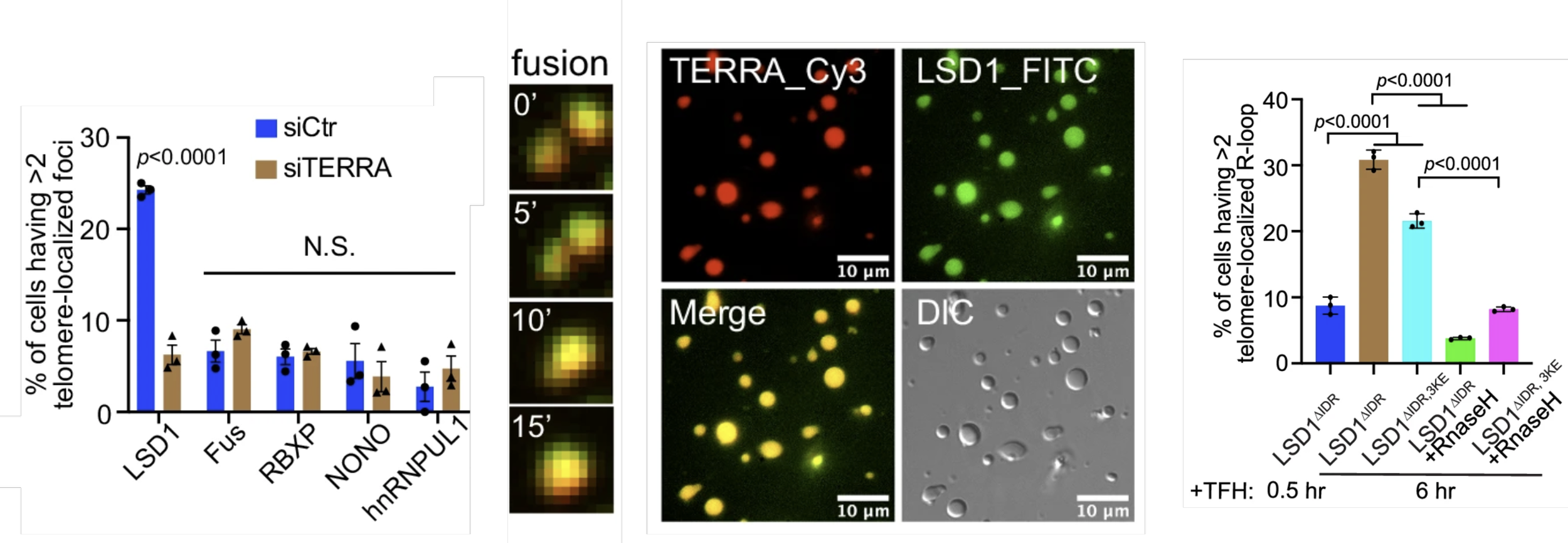
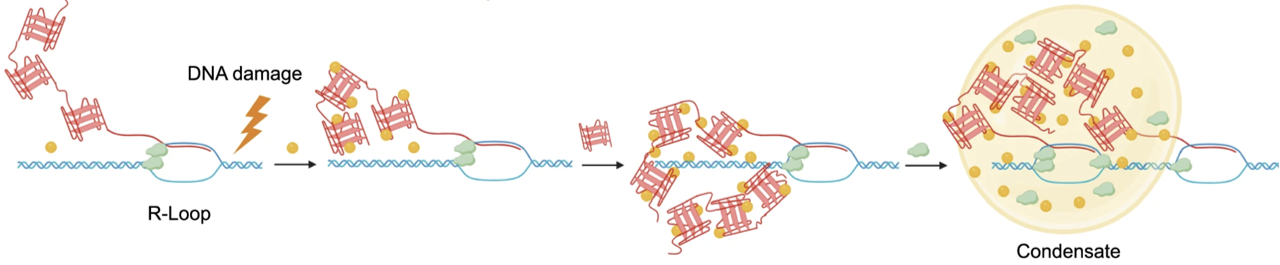
3. M. Xu, D. Senanayaka, R. Zhao, T. Chigumira, A. Tripathi, J Tones, R.M. Lackner, A. R. Wondisford, L.N. Moneysmith, A. Hirschi, S. Craig, S. Alishiri, R. J. O’ Sullivan, D. M. Chenoweth, N. J. Reiter, H. Zhang* “TERRA-LSD1 phase separation promotes R-loop formation for telomere maintenance in ALT cancer cells.” Nature Communications, 2024, 15, 2165-2184.
4. D. Zeng, A. Abzhanova, B. P. Brown, and N. J. Reiter (2021) “Dissecting Monomer-Dimer Equilibrium of an RNase P protein provides insight into the synergistic flexibility of 5’ Leader pre-tRNA recognition.” Front. in Molecular Biosciences, 8: 730274 – 730285
5. 2. A. Abzhanova, A. Hirschi, and N.J. Reiter (2021) “An Exon-Biased Biophysical Approach And NMR Spectroscopy Define The Secondary Structure Of A Conserved Helical Element Within The HOTAIR Long Non-Coding RNA”, J. of Structural Biology, 213 (2): 107728 – 107735
6. N. Schneider, L.J. Tassoulas, D. Zeng, A. Laseke, N.J. Reiter, L.P. Wackett, M St. Maurice (2020) “Solving the conundrum: Widespread proteins annotated for urea metabolism in bacteria are carboxyguanidine deiminases mediating nitrogen assimilation from guanidine.” ACS Biochemistry, 59 (35): 3258-3270
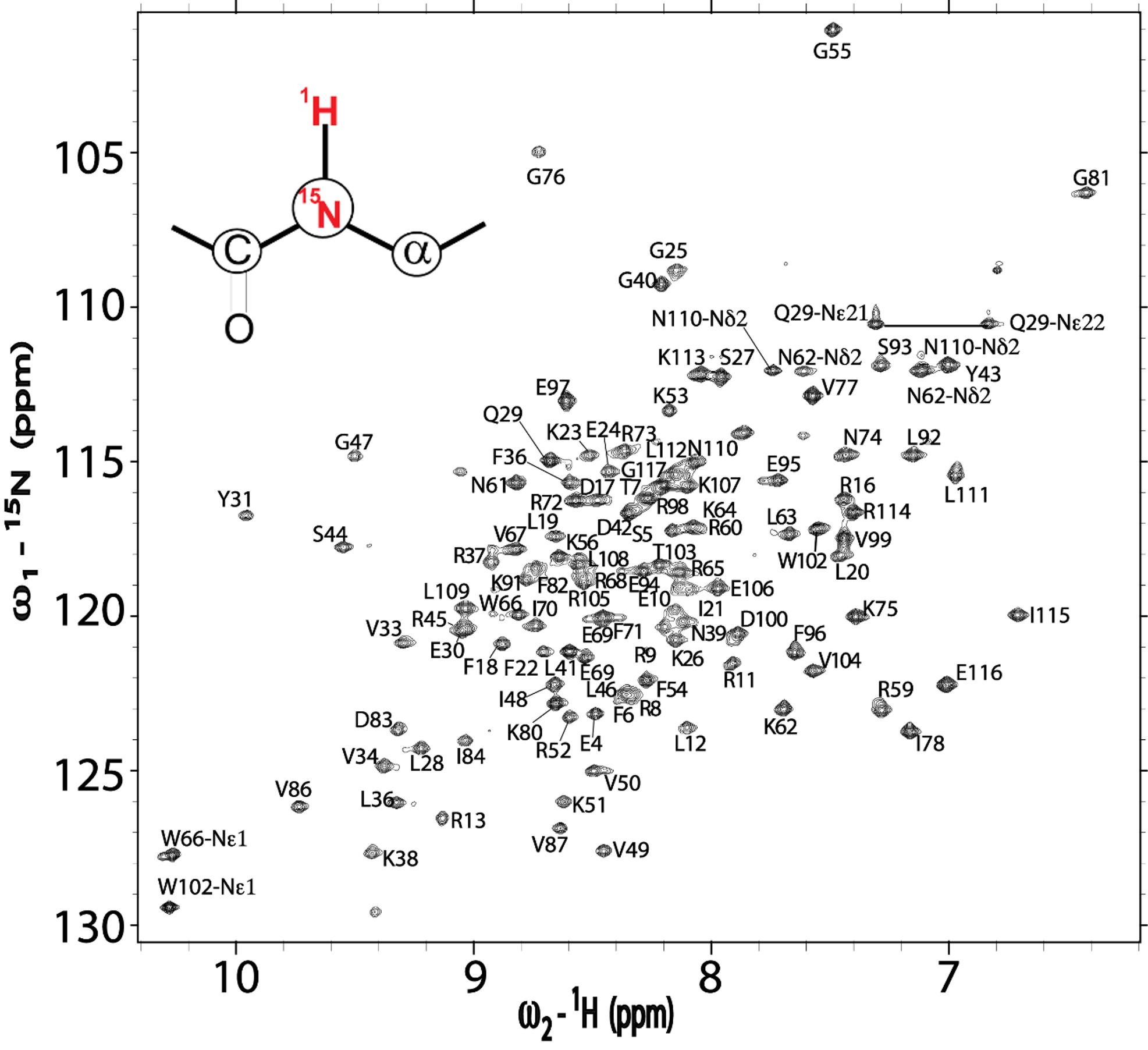
7. D. Zeng, B. P. Brown, M. V. Voehler, S. Cai, N. J. Reiter (2018) “NMR resonance assignments of RNase P protein from Thermotoga maritima.” Biomolecular NMR Assignment, 12 (1): 183-87.
8. W. J. Martin, N.J. Reiter (2017) “Structural Roles of Noncoding RNAs in the Heart of Enzymatic Complexes.” ACS Biochemistry, 56 (1): 3-13
9. A. Hirschi, W.J. Martin, Z. Luka, L.V. Loukachevitch, N.J. Reiter (2016) “G-quadruplex RNA binding and recognition by the lysine specific demethylase-1 enzyme.” RNA, 22 (8)
10. S.E. Collier, M.W. Voehler, D. Peng, R. Ohi, K.L. Gould, N.J. Reiter, and M.D. Ohi (2014) “Structural and functional insights into the N-terminus of Schizosaccharomyces pombe Cdc5.” ACS Biochemistry, 53(41): 6439-51.
11. N.J. Reiter, A Osterman, and A. Mondragón (2012) “The ribonuclease P holoenzyme requires specific, conserved residues for efficient catalysis and substrate positioning.” Nucleic Acids Research, 40(20): 10384-93.
——————————————————————————
12. N.J. Reiter, C.W. Chan, A Mondragón (2011) “Emerging structural themes in large RNA molecules.” Current Opinion in Structural Biology, 21(3): 319-26.
13. J.R. Mohrig, N.J. Reiter, R Kirk, M.R. Zawadski, N Lamarre-Vincent. (2011) “Effect of buffer general acid-base catalysis on the stereoselectivity of ester and thioester H/D exchange in D2O.” Journal of American Chemical Society (JACS), 133(13):5124-8.
14. N.J. Reiter, A Osterman, A Torres-Larios, K.K. Swinger, T Pan, and A. Mondragón (2010) “Structure of a bacterial RNase P holoenzyme in complex with tRNA.” Nature, 468: 784-789.
Comments in: B. Masquida & E. Westhof, RNA 12 (3), 2011. ; P. Schimmel, Nat Rev Mol Cell Biol. 17 (9) 2011. ; T. L. Sheppard, Nat Chem. Biol. 7:2-3, 2011.
15. S Martin-Tumasz, N.J. Reiter, D.A. Brow, S.E. Butcher. (2010) “Structure and functional implications of a complex containing a segment of U6 RNA bound by a domain of Prp24.” RNA, 16(4):792-804.
16. R.D. Sherada, N.J. Reiter, S.E. Butcher, J.L. Keck (2009) “Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB's C terminus.” Journal of Molecular Biology (JMB), 386(3):612-25.
17. N.J. Reiter, L.J. Maher 3rd, S.E. Butcher* (2008) “DNA mimicry by a high-affinity anti-NF-kB RNA aptamer.” Nucleic Acids Research, 36(4):1227-36.
18. E Bae, N.J. Reiter, C.A. Bingman, S.S. Kwan, D Lee, G.N. Phillips, Jr., S.E. Butcher, D.A. Brow. (2007) “Structure and interactions of the first three RNA recognition motifs of splicing factor Prp24.” JMB, 367(5): 1447-58.
19. N.J. Reiter, D Lee, Y.X. Wang, M Tonelli, A Bahrami, C.C. Cornilescu, S.E. Butcher. (2006) “Resonance assignments for the two N-terminal RNA recognition motifs (RRM) of the S. cerevisiae Pre-mRNA processing protein Prp24” Journal of Biomolecular NMR, 36 Suppl 5:58.
20. H Blad, N.J. Reiter, F Abildgaard, J.L. Markley, S.E. Butcher. (2005) “Dynamics and Metal Ion Binding in the U6 RNA intramolecular stem loop as Analyzed by NMR.” Journal of Molecular Biology (JMB), 353(3): 540-55.
21. N.J. Reiter, H Blad, F Abildgaard, S.E. Butcher (2004) “Dynamics in the U6 RNA intramolecular stem loop: A base flipping conformational change.” Biochemistry, 43(43): 13739-47.
22. N.J. Reiter, L.J. Nikstad, A.M. Allman, R.J. Johnson, S.E. Butcher (2003) “Structure of the U6 RNA intramolecular stem-loop harboring an S(P)-phosphorothioate modification.” RNA, 9(5): 533-42.
23. T A Reiter, N J Reiter, F Rusnak. (2002) “Manganese is a native metal ion activator for bacteriophage lambda protein phosphatase.” Biochemistry, 41(51): 15404-9.
24. N J Reiter, D J White, F Rusnak (2002) “Inhibition of Bacteriophage lambda Protein Phosphatase by Organic and Oxoanion Inhibitors.” Biochemistry, 41(3): 1051-9.
25. D J White, N J Reiter, R A Sikkink, L Yu, F Rusnak (2001) “Identification of the high affinity Mn2+ binding site of bacteriophage lambda phosphoprotein phosphatase: Effects of metal ligand mutations on electron paramagnetic resonance spectra and phosphatase activities.” Biochemistry, 40(30): 8918-8929.
26. W C Voegtli, D J White, N J Reiter, F Rusnak, A C Rosenzweig (2000) “Structure of the bacteriophage lambda Ser/Thr protein phosphatase with Sulfate Ion Bound in Two Coordination Modes.” Biochemistry, 39(50): 15365-15374.
Senanayaka et. al, JBC(2024)
Xu et. al, Nature Communications (2024)
Zeng et. al, Bio. NMR assignment (2020)
Zeng et. al, Front. in Mol. Bio. (2021)
Hirschi et. al, RNA. (2016)
Schneider et. al, ACS Biochemistry. (2020)
Abzhanova et. al, J. Struct. Biol. (2021)
Hirschi et. al, RNA. (2016)
Martin & Reiter, ACS Biochemistry (2017)
Image adapted from: Fig 5D, Lan et. al, Science (2020)