Human Histone Lysine Specific Demethylase
-
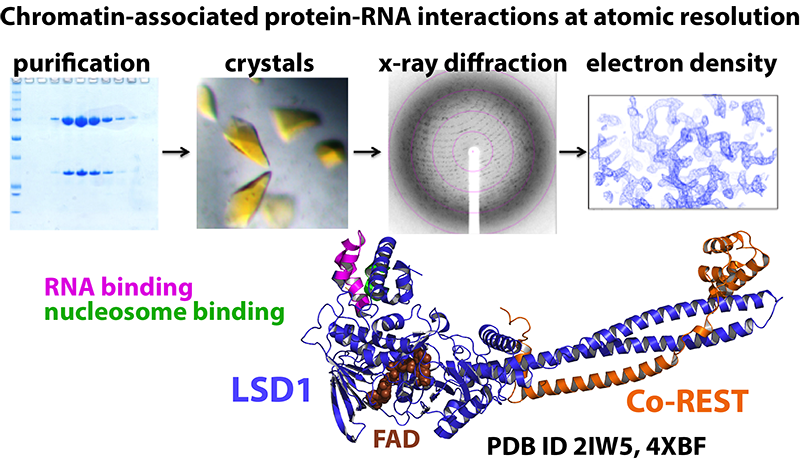
This NIH sponsored project examines the protein/RNA-mediated mechanisms of the lysine specific histone demethylase-1 (LSD1 or KDM1A) enzyme. LSD1 is a flavin-dependent amine oxidase that plays key roles in embryogenesis and misregulated LSD1 correlates with cancer progression. Human LSD1 is considered an epigenetic “reprogramming” enzyme because it functions to remove methyl groups from lysine position 4 and 9 of the critical histone 3 protein. Thus, LSD1 functions as a demethylation enzyme, and it can interact with over 100 different proteins, the nucleosome, various RNAs, and small metabolites. Due to LSD1’s diverse gene regulatory network, novel ways to probe LSD1 in a pathway-specific manner has high therapeutic value. We use RNA-LSD1 and disease related intrinsic disorder protein (IDP) - LSD1 complexes as model systems to understand the myriad of interactions and functions of chromatin-associated proteins. To this end, we seek to use this information to develop improved therapeutics to selectively target LSD1-mediated cancer progression.
Bacterial Ribonuclease P (RNase P) structure and function
-
We are interested in the structure and conformational dynamics of large catalytic RNA molecules (ribozymes). One RNA-protein complex, termed Ribonuclease P (RNase P), is involved in tRNA biogenesis and is considered an essential ribozyme that is found in all 3 domains of life (bacteria, archaea, and eukaryotes). We use RNase P as a model to understand how large RNAs and RNA binding proteins contribute to catalysis. Because RNase P represents a nearly universal processing event that is linked to the origins of the genetic code, we seek to understand its conformational diversity at the level of tertiary structure. The discovery that the RNA component of RNase P is directly involved in catalysis demonstrates that RNA, just like proteins, is a highly versatile biomolecule and can assemble into a complex three-dimensional architecture. RNase P serves as an ideal system to explore how proteins assemble and interact with large catalytic RNAs to carry out a specific biological function. For this project, we integrate biochemistry and biophysical methods, focusing on the conformational dynamics of RNase P at atomic resolution.